Sorin Lab News
2022/23 Updates (see archive)
 Biochemist Daniel Chavez-Bonilla received the 2023 John H. Stern Summer Research Assistantship and Award in Physical Chemistry
Biochemist Daniel Chavez-Bonilla received the 2023 John H. Stern Summer Research Assistantship and Award in Physical Chemistry
 Computer Science major and Sorin Lab SysAdmin Nicholas Tsimerekis earned a 2023 Summer Internship @ NASA Jet Propulsion Labratory!
Computer Science major and Sorin Lab SysAdmin Nicholas Tsimerekis earned a 2023 Summer Internship @ NASA Jet Propulsion Labratory!
 Sorin Labber Karel Aceituno will start the Neuroscience PhD program at USC in the fall 2023 semester
Sorin Labber Karel Aceituno will start the Neuroscience PhD program at USC in the fall 2023 semester
 Information Systems major Daniel Sanchez was admitted to the prestigious McNair Scholars program!
Information Systems major Daniel Sanchez was admitted to the prestigious McNair Scholars program!
2021 Updates (see archive)
 Undergrad lab members Jia Mao & Danna De Boer both received CSULB Summer Research Awards to support their summer research efforts
Undergrad lab members Jia Mao & Danna De Boer both received CSULB Summer Research Awards to support their summer research efforts
 Biochemistry major Karel Aceituno was awarded the 2021 John H. Stern Summer Research Assitantship & admitted to the RISE program!
Biochemistry major Karel Aceituno was awarded the 2021 John H. Stern Summer Research Assitantship & admitted to the RISE program!
 BME major and Sorin Lab member Jessica Moore has accepted admission to the UC Irvine Biomedical Engineering Ph.D. program for fall 2021
BME major and Sorin Lab member Jessica Moore has accepted admission to the UC Irvine Biomedical Engineering Ph.D. program for fall 2021
 Chem/German double major Danna De Boer was admitted to a research internship in Germany where she'll model gecko surface adhesion
Chem/German double major Danna De Boer was admitted to a research internship in Germany where she'll model gecko surface adhesion
 ChemEng major Nguyet Nguyen earned a Best Poster award @ the Spring ACS Meeting and a SoCalGas research assistantship
ChemEng major Nguyet Nguyen earned a Best Poster award @ the Spring ACS Meeting and a SoCalGas research assistantship
2020 Updates (see archive)
 BS Chemistry major Daniel Figueroa was awarded the 2020 Stern Award for Excellence in Physical Chemistry
BS Chemistry major Daniel Figueroa was awarded the 2020 Stern Award for Excellence in Physical Chemistry
 Chemical Engineering major Jia Mao was awarded a Women & Philanthropy Scholarship to support his research into BChE inhibition
Chemical Engineering major Jia Mao was awarded a Women & Philanthropy Scholarship to support his research into BChE inhibition
 Congratulations to lab alumnus and University of Chicago Biophysics PhD student Walter Alvarado on becoming a NASA Fellow!
Congratulations to lab alumnus and University of Chicago Biophysics PhD student Walter Alvarado on becoming a NASA Fellow!
Computational Biochemistry & Biophysics
Members of the Sorin Lab study the structure and dynamics of biological molecules via computer modeling and simulation.
Using cluster computing and the Folding@Home Distributed Computing Project, we can apply a variety of models and
methods to sample large numbers of independent simulations: we are now capable of simulating some molecular events
in dynamic equilibrium, making simulation a powerful tool with which to probe topics in molecular biology and biophysics
including molecular recognition and drug design, RNA and protein folding, and related processes.
[ Click here to apply for a position (.doc application) ]

Front: Ben Doan, Alexia Chinn, Daniel Chavez-Bonilla, Carlos Villalobos, and Megan Calderon
Recent Publications
 |
A Hands-On Collaboration-Ready Single- or Interdisciplinary Computational Exercise in Molecular Recognition and Drug Design.
Patrick Allen, Nguyet Nguyen, Nicholas D. Humphrey, Jia Mao, Daniel Chavez-Bonilla, and Eric J. Sorin
(2024).
Education Sciences, 14, 139, doi.org/10.3390/educsci14020139.
|
 |
Energetically unfavorable protein angles: Exploration of a conserved dihedral angle in triosephosphateisomerase.
Patrick W. Allen, Jordan A. Cook, Anh N. Colquhoun, Eric J. Sorin, Enrico Tapavicza, and Jason P. Schwans
(2022).
Biopolymers, 113, e23525, doi.org/10.1002/bip.23525.
|
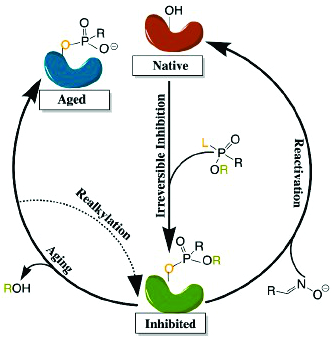 |
A Comprehensive Review of Cholinesterase Modeling and Simulation.
Danna De Boer†, Nguyet Nguyen†, Jia Mao, Jessica Moore, & Eric J. Sorin* (2021).
Biomolecules, 11, 580, DOI:10.3390/biom11040580.
|
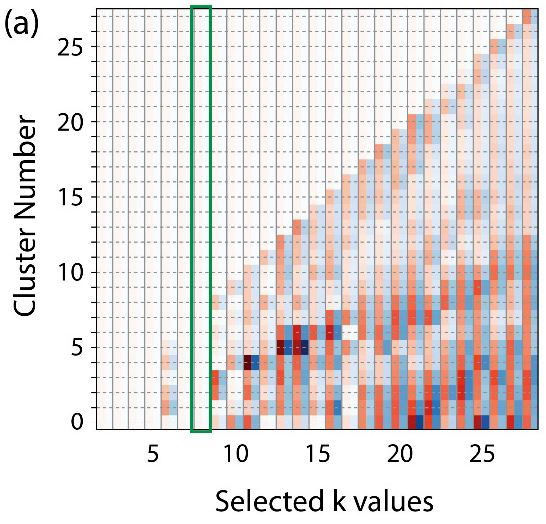 |
Overcoming the Heuristic Nature of k-means Clustering: Identification and Characterization of Binding
Modes from Simulations of Molecular Recognition Complexes. Parker Ladd Bremer, Danna De Boer, Walter Alvarado, Xavier Martinez, & Eric J. Sorin* (2020).
Journal of Chemical Information and Modeling, 60(6), 3081 - 3092, DOI:10.1021/acs.jcim.9b01137.
|
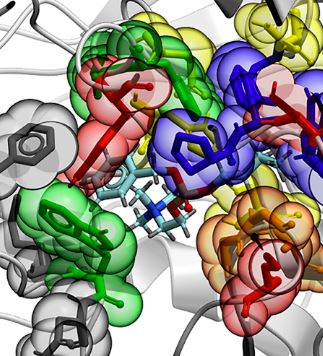 |
Understanding the Enzyme-Ligand Complex: Insights from All-Atom Simulations of Butyrylcholinesterase Inhibition. Walter
Alvarado, Parker Ladd Bremer, Angela Choy, Helen N. Dinh,
Aingty Eung, Jeannette Gonzalez, Phillippe Ly, Trina Tran,
Kensaku Nakayama*, Jason P. Schwans*, and Eric J. Sorin* (2019).
Journal of Biomolecular Structure & Dynamics, 38(4), doi:DOI:10.1080/07391102.2019.1596836.
|
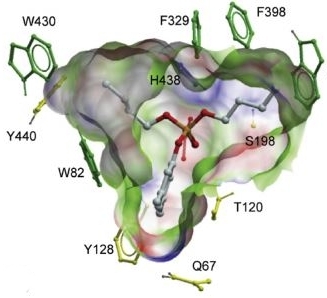 |
Synthesis, biochemical evaluation, and molecular modeling studies of aryl and arylalkyl di-n-butyl phosphates,
effective butyrylcho linesterase inhibitors. Kensaku Nakayamaa*, Jason P. Schwans*, Eric J. Sorin*, Trina Tran,
Jeannette Gonzalez, Elvis Arteaga, Sean McCoy & Walter Alvarado
(2017).
Bioorganic & Medicinal Chemistry, 25(12), 3171–3181, doi:10.1016/j.bmc.2017.04.002.
|
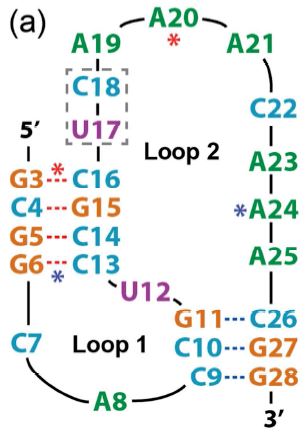 |
Ensemble simulations: folding, unfolding and misfolding of a high-efficiency frameshifting RNA pseudoknot
. Khai K. Q. Nguyen, Yessica K. Gomez, Mona Bakhom, Amethyst Radcliffe, Phuc La, Dakota Rochelle, Ji Won Lee & Eric J. Sorin*
(2017). Nucleic Acids Research 45, doi:10.1093/nar/gkx012.
|
